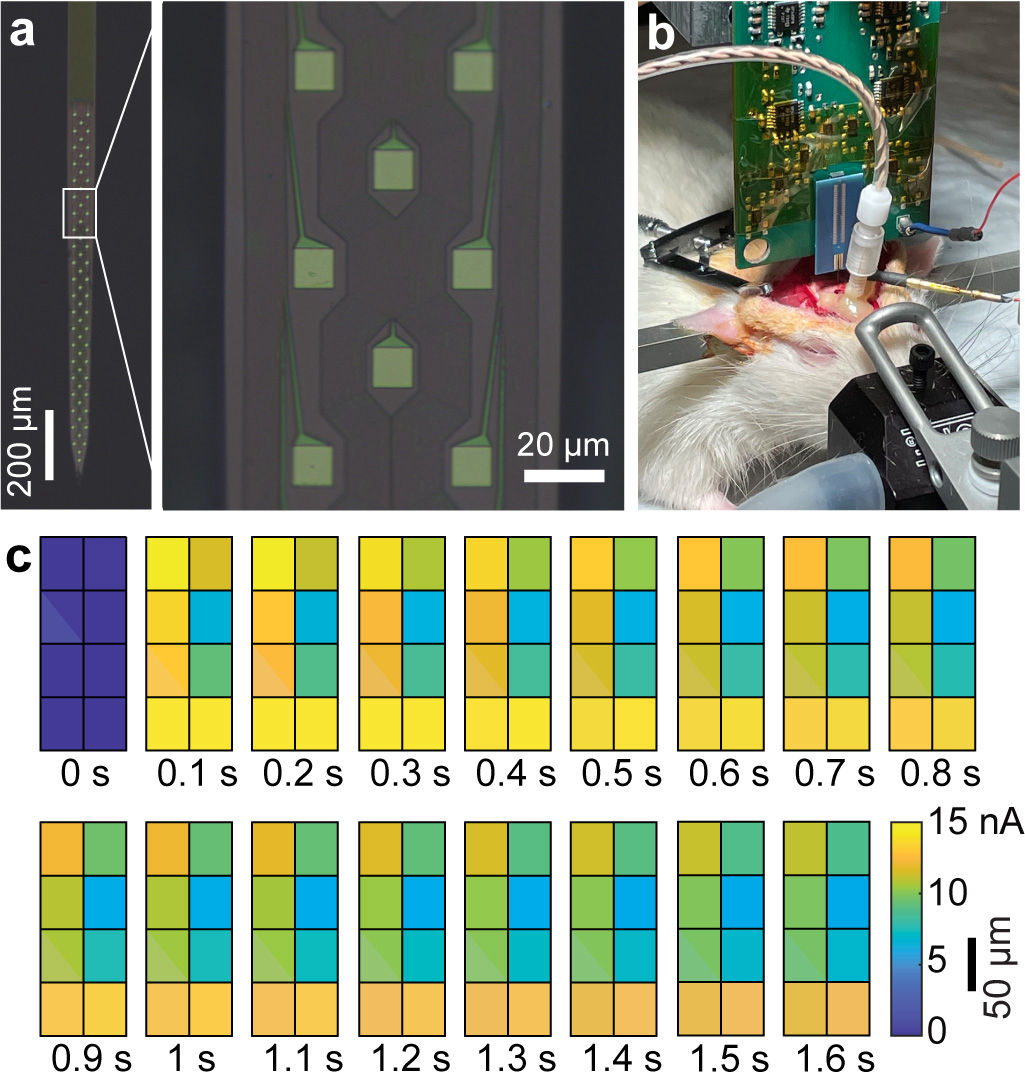
Biomedical Circuits and Systems
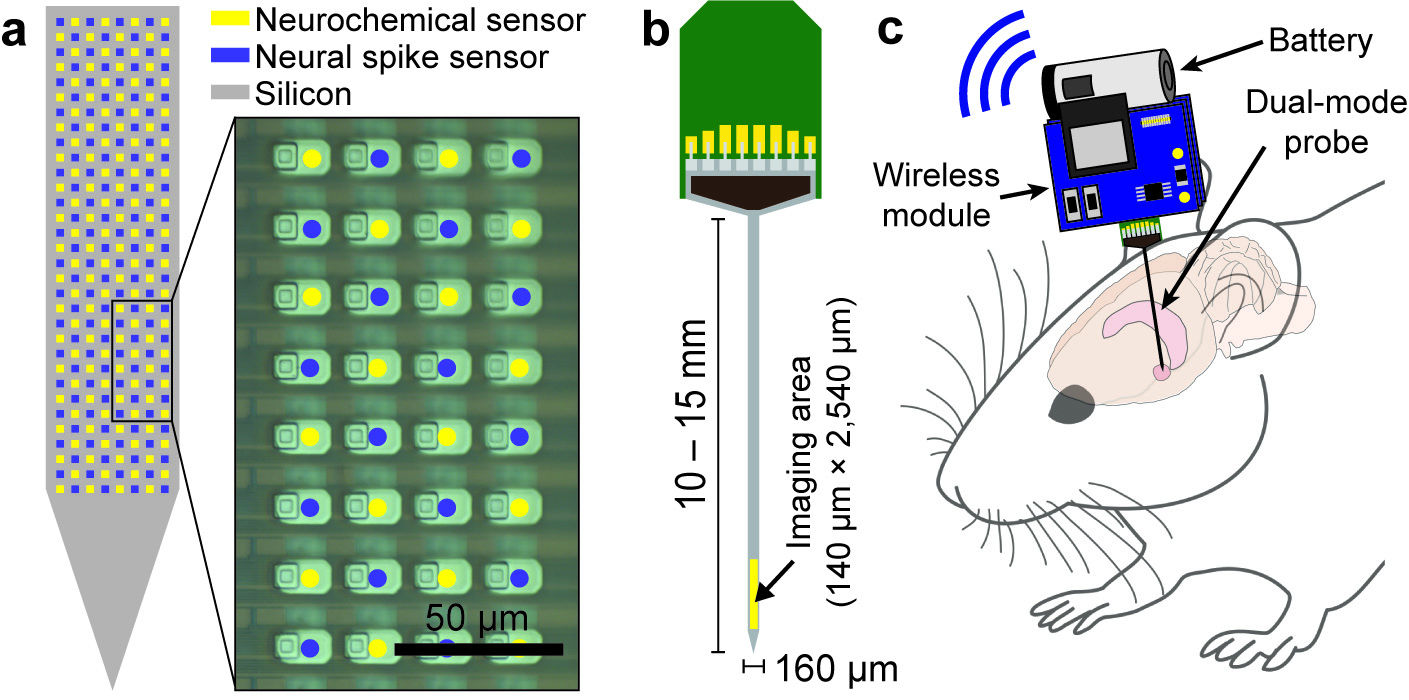
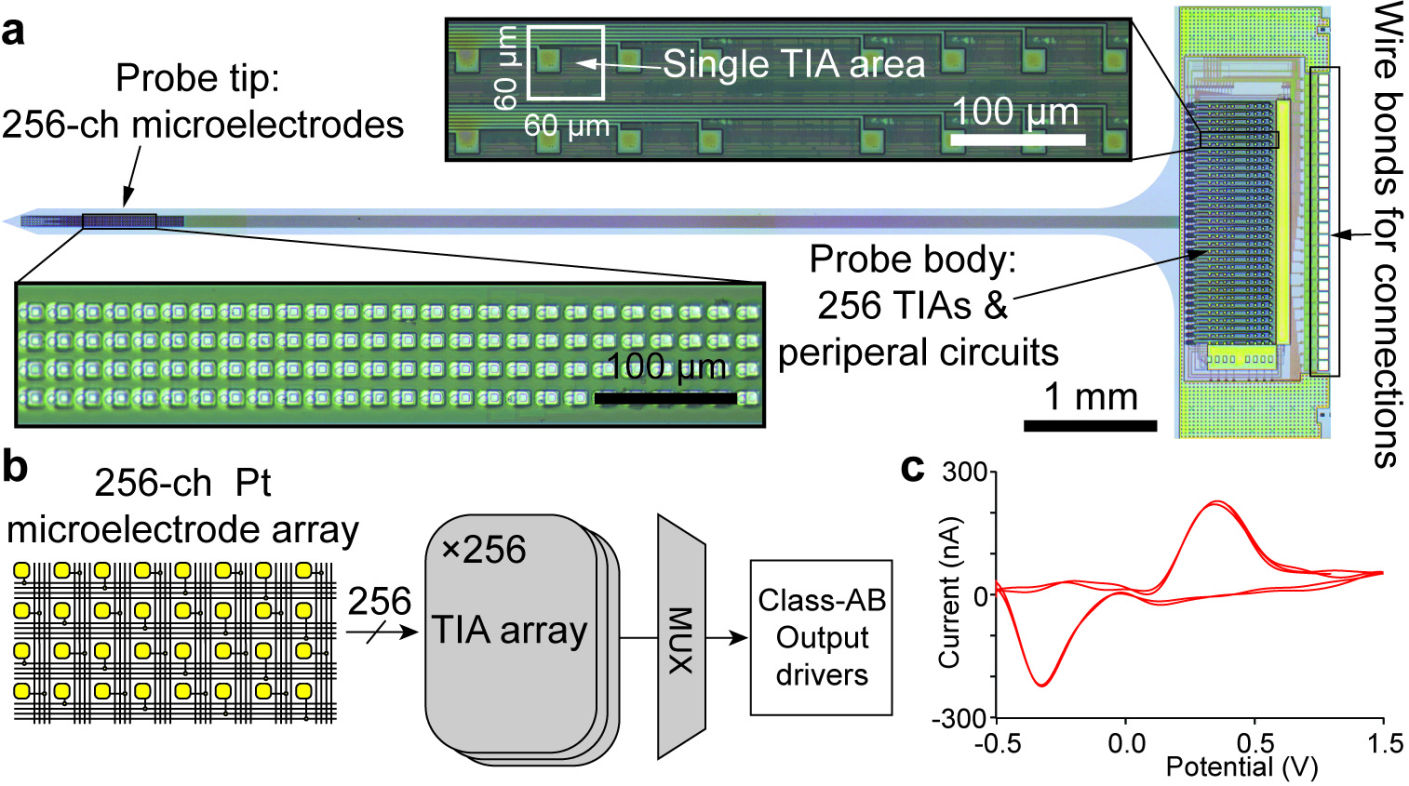
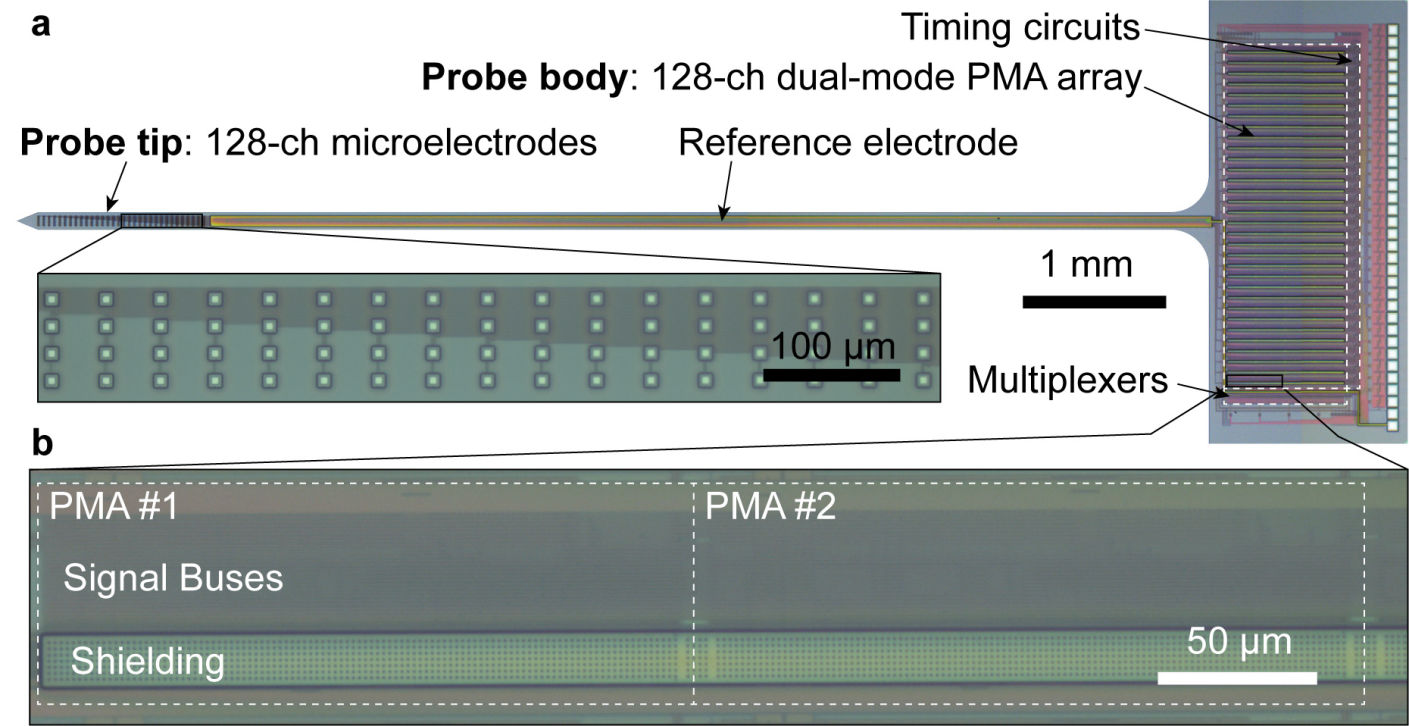
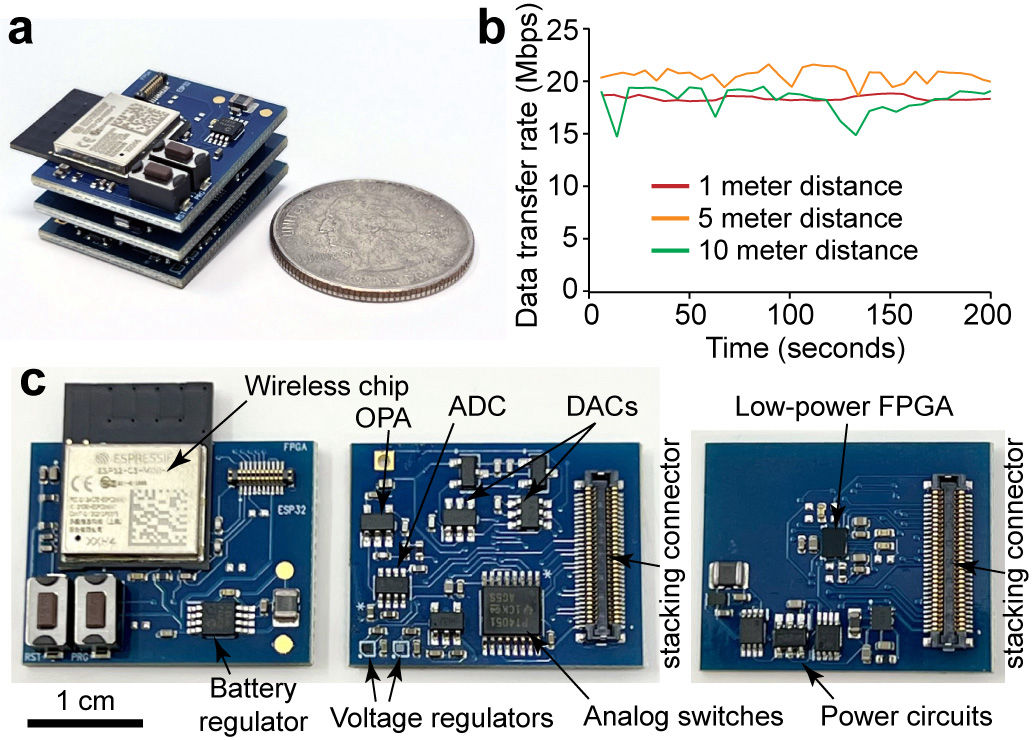
Our research group develops biomedical circuits and systems including electrodes, IC chips, PCB, FPGA, data acquisition systems, firmware and software! The following pictures are a few examples of the chips that we've developed for both in vitro and in vivo applications
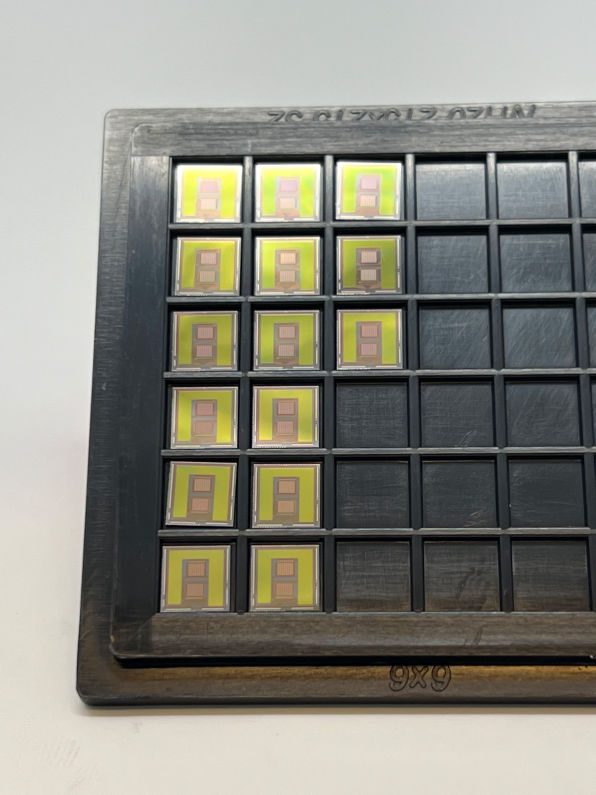
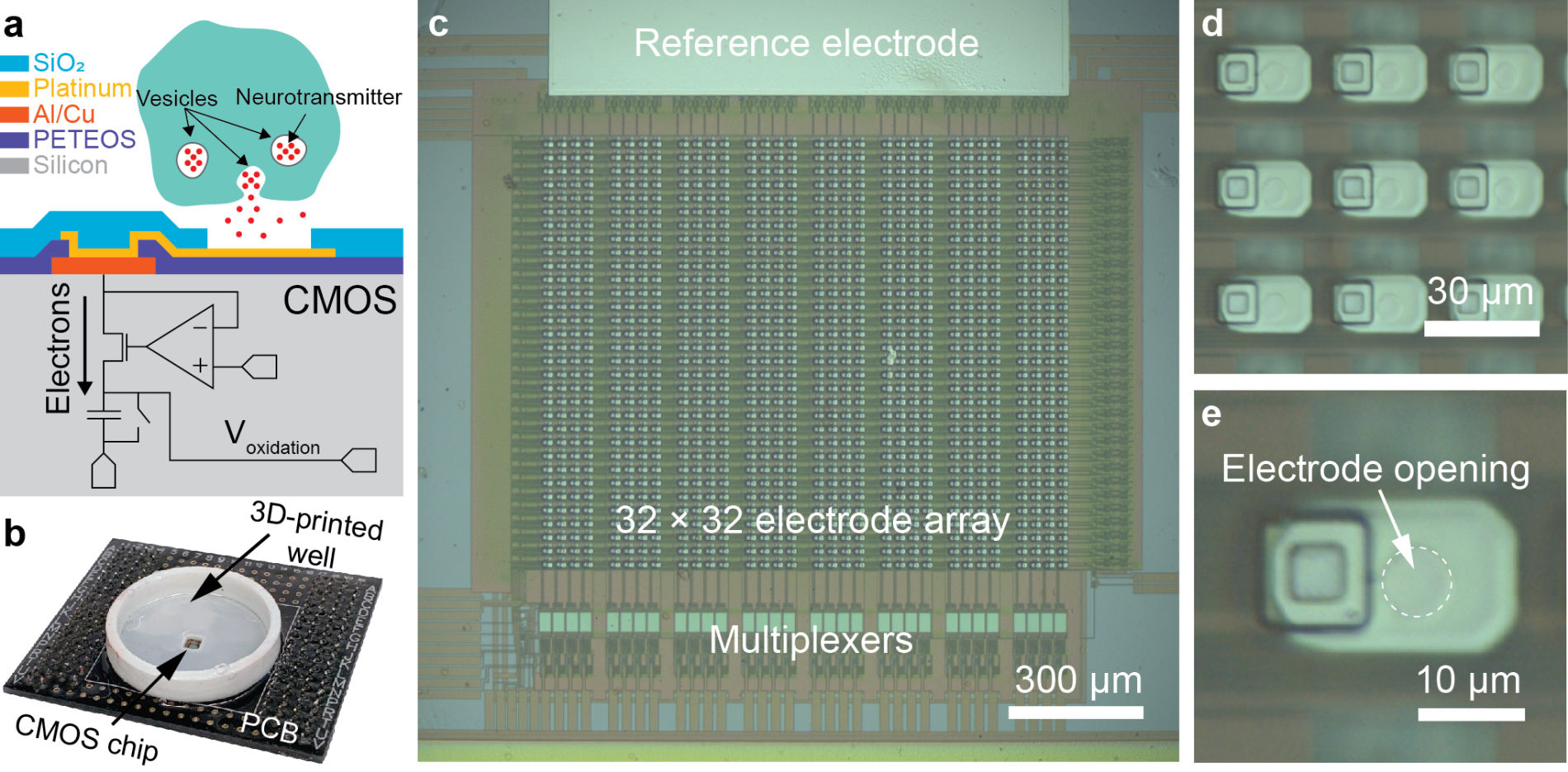
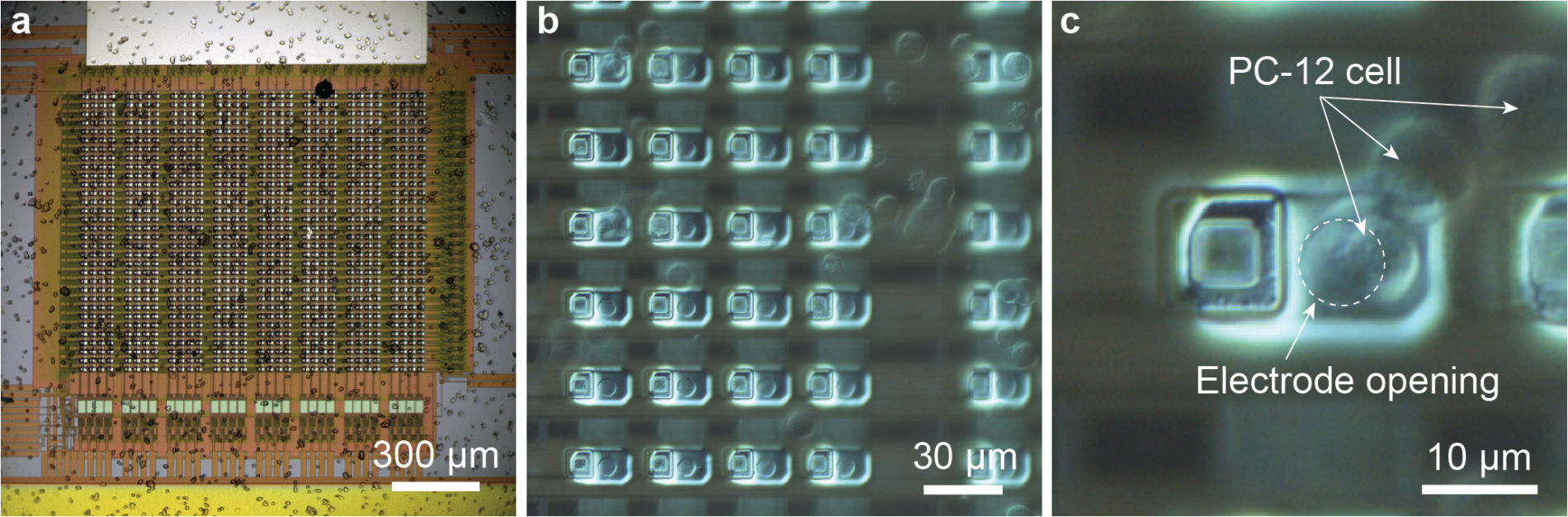
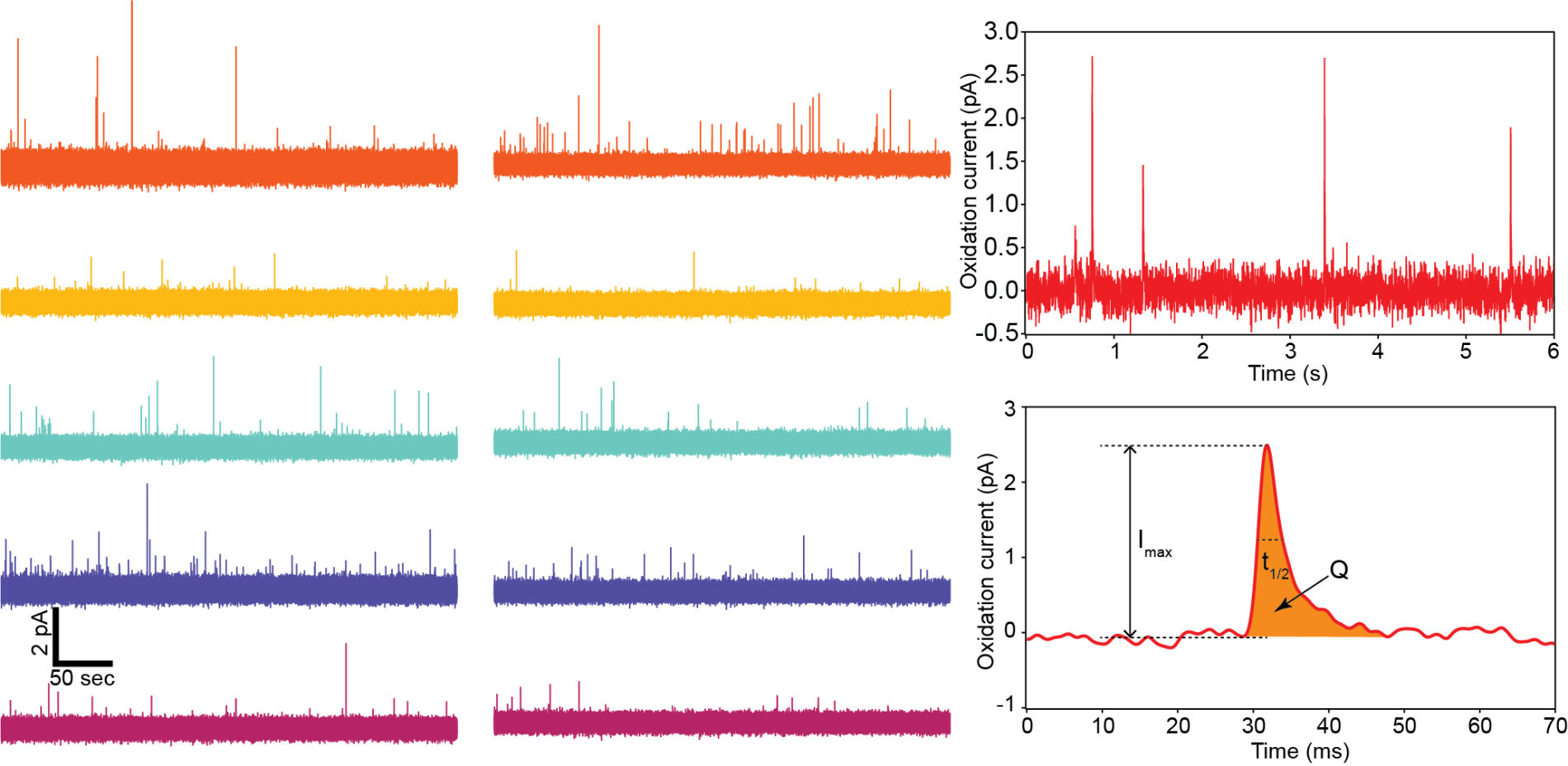
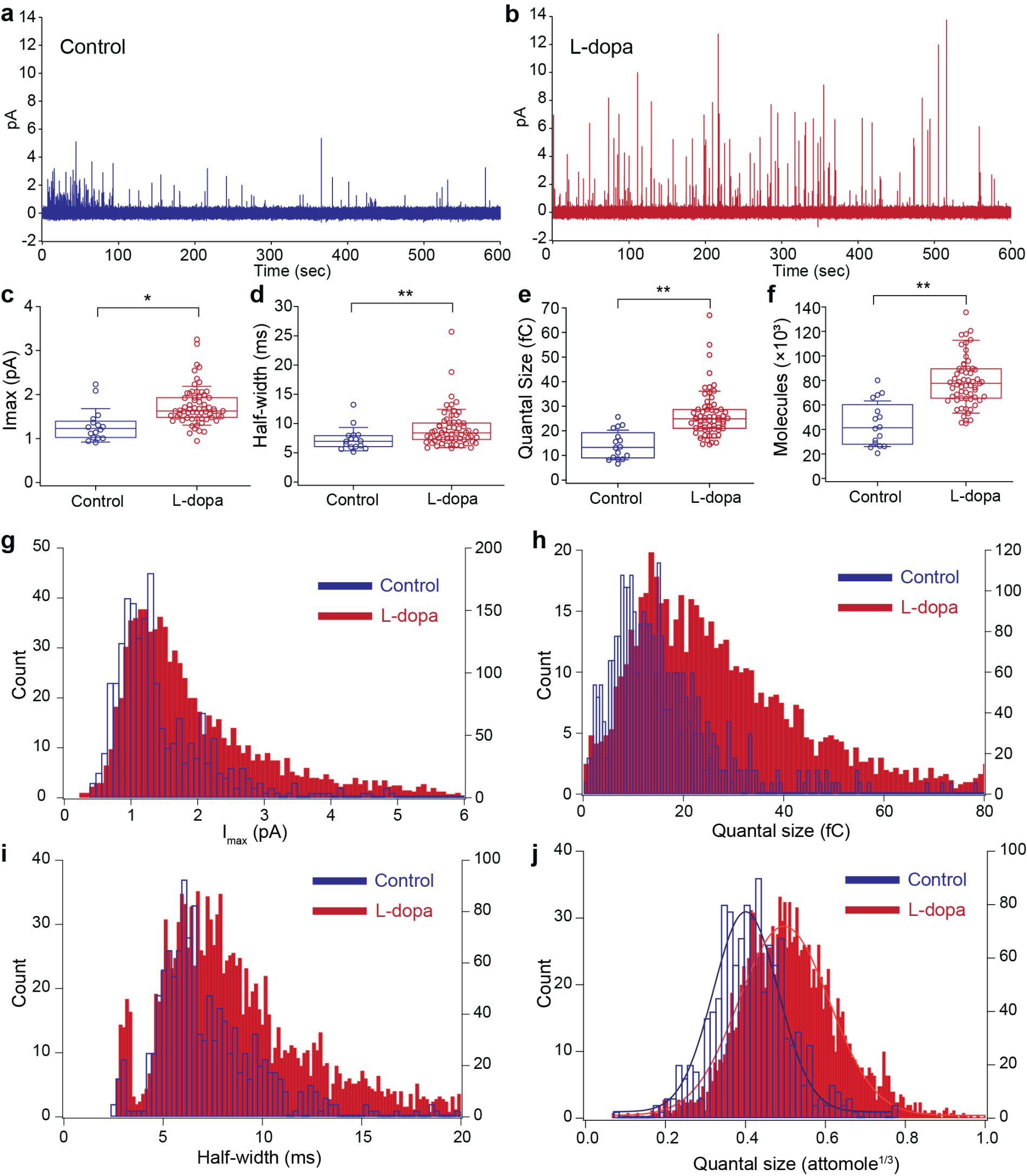
1000-ch in vitro neurochemical sensor (integrated 1024-ch on-chip electrodes + 1024 TIA amplifiers)

1000-ch in vitro neurochemical sensor with FSCV (integrated 1024-ch on-chip electrodes + 1024 TIA amplifiers)

256-ch fully wireless BMI chip (integrated power coil + amplifier + electrodes)